The ubiquitin-proteasome pathway (UPP) is one of the major destruction ways to control the activities of different proteins. The function of UPP is to eliminate dysfunctional/misfolded proteins via the proteasome, and these specific functions enable the UPP to regulate protein quality in cells. Therefore defects in UPP are expected to disturb cellular homeostasis and are detrimental to cell survival.
The steps of Ubiquitin-Proteasome Pathway
The UPP is the principal negative regulatory mechanism proteins which are post-translationally marked with ubiquitin (Ub) or multiple ubiquitin molecules attached to a lysine side chain on a protein. The Ub provides a recognition signal for the 26S proteasome, and protein modified with Ub, shuttling factors facilitate its transport to the 26S proteasome where dedicated receptors initiate its degradation. It has developed an ingenious way to divide this complex biochemical machinery into two discrete and successive steps:
- Firstly, step A is a specific recognition process, employing the Ub conjugation cascade that uses a highly modular approach in different combinations for different purposes. Specifically, three different sets of enzymes (i.e. E1, E2, and E3) shuttle Ub and eventually link Ub to the protein substrate. Different combinations of E2 or E3 or both recognize each substrate’s unique degradation signal, thereby conferring exquisite specificity for ubiquitination of diverse protein substrates.
- The step B, degradation of the tagged substrate by the 26S proteasome is an indiscriminate destruction process, mediated by the proteolytic proteasome core. This indiscriminate proteolytic step provides directionality for a signaling pathway, i.e., once a protein is committed for degradation, there is no return, ensuring that partially degraded proteins do not interfere with biological processes. Ultimately, most ubiquitinated proteins are recognized by the 26S proteasome, unfolded, and threaded into the 20S proteolytic chamber in an ATP-dependent manner.
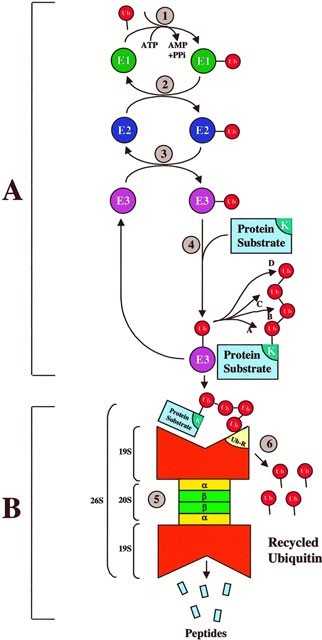
Figure 1. The steps of Ubiquitin-Proteasome Pathway. (Ciechanover, A. 1998)
Process and Regulation of Ubiquitin-Proteasome Pathway
Ubiquitin is a 9 kDa regulatory protein which attaches to substrate proteins to form a post-translational modification, ubiquitination. The long poly-ubiquitin chains are targeted for the degradation of the 26S proteasome. The 26S proteasome, composed of 19S subunits to either side of the 20S subunit, is the proteolytic machinery associated with the ubiquitin pathway. It should be emphasized that the specificity of ubiquitination is largely determined by a series of E3 enzymes and E3 multiprotein complexes, each of which is specific to one or a few corresponding protein substrate(s) and of E2 enzymes, each of which is dedicated to their cognate E3 enzyme(s). As a result, different combinations of E2 and E3 enzymes allow selective tagging and degradation of specific intracellular proteins. In contrast, there is a single family of E1 that is highly conserved.
The protein Ubc13 is a member of the Ub-conjugating (E2) enzyme family that receives Ub from E1 enzyme. Once the protein substrate is mono-ubiquitinated, a poly-ubiquitin chain is formed through the same ubiquitination conjugation cascade, in which the carboxyl group of the carboxy-terminal Gly-76 of ubiquitin is covalently linked to an internal Lys residue of ubiquitin that is already conjugated to the protein substrate. TNFα induces receptor-interacting protein kinase (RIP) poly-ubiquitination, and ubiquitinated RIP associates with NEMO (NF-κB essential modulator) to activate inhibitors of nuclear factor kappa B (IκB) kinase complex which is formed by IKKα and IKKβ. This results in the ubiquitination of IκB, and ubiquitinated IκB is separated from NF-κB and directed to the 26S proteasome, which is facilitated by valosin-containing peptide (VCP). NF-κB is released to the nucleus to activate the transcription of target genes.
The binding of the ligand Ectodysplasin (EDA-A1) to the TNF-receptor EDAR results in the formation of a complex containing EDAR ADD, TRAF6, TAB2, and TAK1. TAK1 can also activate the IKK, which in turn phosphorylates IKK.
Downregulation of NF-κB activation by the T-cell receptor (TCR) involves the TCR-induced ubiquitination and degradation of Bcl10. Bcl10 can stimulate K63 poly-ubiquitination of NEMO, and TRAF6 can catalyze mono-ubiquitination of NEMO or K63 poly-ubiquitination of itself. Furthermore, K63 poly-ubiquitination of RIP is recognized by NEMO as a binding and activation signal.
Many ubiquitin-like proteins (UBLs), such as SUMO, NEDD8 also have novel functions and influence diverse biological processes. SUMO modification often acts antagonistically to that of ubiquitination and serves to stabilize protein substrates. Proteins conjugated to UBLs are typically not targeted for degradation by the proteasome, but rather function in diverse regulatory activities. Attachment of UBLs might alter substrate conformation, affect the affinity for ligands or other interacting molecules, alter substrate localization and influence protein stability.
Ubiquitin-Proteasome Pathway in Pathologies
The important role of UPP was awarded the Nobel Prize in Chemistry in 2004. It has emerged as a central player in the regulation of several diverse cellular processes, affecting DNA transcription, cell cycle, inflammation, ribosome biogenesis and soon. Indeed, defects in a various component of UPP lead to several human diseases, including cancer and neurodegenerative diseases, rendering pharmacological modulation of this pathway a promising future therapeutic strategy.
References:
- Ciechanover, A. The ubiquitin–proteasome pathway: on protein death and cell life. The EMBO Journal.1998, 17(24): 7151-7160.
- Herrmann, J; et al. Ubiquitin and ubiquitin-like proteins in protein regulation. Circulation Research. 2007, 100: 1276-1291